Abstract
Background: Bone marrow microenvironment plays a crucial role in the pathogenesis of multiple myeloma (MM) by supporting plasma cell growth, survival and drug resistance. These abilities have been attributed in part to the secretion of growth factors and cytokines by bone marrow mesenchymal stromal cells (BM-MSC). microRNAs (miRNAs) are promising prognostic biomarkers and therapeutic targets that can be released and transferred among MM cells and BM-MSCs as cell-cell communication. Recently, we defined a serum 14-miRNA signature which was associated with progression and complete remission (CR) after autologous stem-cell transplantation (ASCT) in patients with MM. Patients in CR showed a partial recovery of the normal serum miRNA levels and these are similar to those samples from patients with monoclonal gammopathy of undetermined significance (MGUS). Thus, these results highlight the potential value of serum miRNAs as diagnostic and prognostic biomarkers with a cell origin different from malignant plasma cells. The aim of this study was to discover whether the MSC miRNA profile varies depending on the status of the disease.
Methods: Mononuclear cells from BM samples were cultured in DMEM containing 10% FBS. After a week, non-adherent cells were removed, whereas BM-MSC were selected by their adherence to the plastic and their phenotype was confirmed by multiparametric flow cytometry. We analyzed 670 microRNAs in 20 primary BM-MSC from patients with MGUS (N=4), symptomatic MM (N=8) and MM in CR (N=8). miRNAs differentially expressed were identified according to a supervised analysis using significance analysis of microarrays (SAM) and Student's t-test based on multivariate permutation (with random variance model). miRNAs differentially expressed between groups of patients were validated in an extended cohort of BM-MSC from patients (N= 82) in all the different status disease: MGUS (N=23), MM at diagnosis (N=13), relapsed MM (N=11), MM in partial response (PR) (N=13) and MM in CR (N=22), as well as in malignant plasma cells (CD38+) from those patients with symptomatic MM. In order to identify miRNA targets, we used the RmiR package to cross-correlate the miRNA expression data from the present study with our findings on the gene expression signature in 12 BM-MSCs from patients (4 MGUS, 4 symptomatic MM and 4 in CR), based on the predicted targets from TargetScan and miRBase databases.
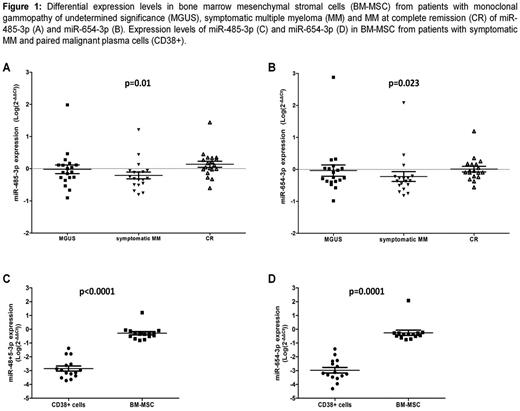
Results: In the screening phase, we identified a miRNA profile of 10 miRNAs (miR-663b, miR-654-3p, miR-206, miR-411*, miR-885-5p, miR-668, miR-638, miR-485-3p, miR-744* and miR-199a) differentially expressed between patients with symptomatic MM and MM in CR (adjusted p-value <0.0001). In the validation phase, miR-485-3p and miR-654-3p resulted differentially expressed in the three groups of patients: MGUS, symptomatic MM and patients in CR (ANOVA test: p=0.0101 and p=0.0228, respectively) (Figure 1A and 1B). The levels of these miRNAs were significantly decreased in patients with MM when compared with MGUS, and these levels seemed to recover when patients achieved CR. These two miRNAs (miR-485-3p and miR-654-3p) were also correlated with all degrees of response in MM: diagnosis, relapse, PR and CR (ANOVA test: p=0.0154 and p=0.0487, respectively). Moreover, paired cross-correlation among these two miRNAs expression with our results in mRNA gene expression profile data showed several potential gene targets with a biological interest, such as MELK and MAP2K2 (correlation index < -0.8). Finally, miR-485-3p and miR-654-3p showed a higher expression in BM-MSC than in MM CD38+ cells (Figure 1C and 1D), suggesting MSC as cell of origin for these miRNAs.
Conclusion: In patients with monoclonal gammopathies, miR-485-3p and miR-654-3p expression in mesenchymal stromal cells from bone marrow is related to the status of the disease and the response to treatment in MM. The modulation of miRNAs profile in the bone marrow microenvironment around malignant plasma cells could be an important mechanism of control by cell-cell communication and the basis for potential biomarkers in serum and bone marrow.
Rosinol: Celgene: Honoraria; Janssen: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal